A substantially updated version of the hypertextbook is available here. Please migrate to that version. This one will eventually disappear.
Objectives
The objectives of this section are to provide you with
an understanding of some of the key characteristics of biofilms;
a recognition of how microorganisms that are part of a biofilm are different from those same microorganisms in isolation.
Outcomes
Upon completion of this section, you will be able
to summarize some of the key characteristics of biofilms;
to discuss some of the ways in which microorganisms which are part of a biofilm differ from the same microorganisms that exist in isolation;
to describe why microorganisms that exist in a biofilm are harder to destroy than the same microorganisms in isolation;
to discuss how microorganisms in a biofilm can communicate with each other.
What are key characteristics of biofilms?
About Section 4
In this section we introduce you to some of the key characteristics of biofilms. When we as scientists and engineers begin to learn about a new life system in order to exploit it for good or to destroy it if it is harmful, we need to understand as much as possible about it. What are its characteristics, and how will this knowledge help us accomplish what we need to do? The more we know about how a system functions, the more we know about how to deal with it. Quite a few things are now known about biofilms, but there is a lot left to uncover. We discuss a few of the things we have learned about biofilms here that help give us insight into why traditional forms of treatment do not seem to work well on biofilms and how we might develop more effective treatments. This overview will prepare you for later modules in which some of these things are explored in depth.
1. Biofilms are complex, dynamic structures
Biofilms are remarkably heterogeneous. Many measurements and observations have been made of various biofilms; they all point to the diversity of individual biofilm colonies. As we have mentioned before, in typical, naturally occurring biofilms (as opposed to some that are grown in a laboratory for experimental reasons) there are nearly always a large number of different kinds of microorganisms living together. In addition, different biofilms seem to exhibit different internal structures, different chemical properties, different electrical properties, and, indeed, different properties of just about any other measurement or observation that can be made. Each of these properties seems to contribute to the characteristics of the biofilm as a whole that make it different (e.g., hard to kill) compared to dealing with each of the microorganisms in isolation (not in a biofilm, but in a planktonic environment).
So here is a big question. If there is such a wide diversity of properties in different biofilms, how can we expect to find characteristics that apply to all biofilms? We are glad you ask. What has been discovered is that in spite of their wide diversity, biofilms do seem to have some common attributes, such as their ability to grow on virtually any surface, how they attach to a surface, their mode of growth, their ability to spread, how they are nourished, how they maintain themselves as a colony, and so forth.
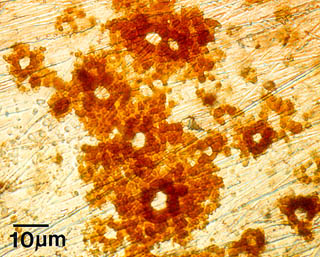
For example, the image at the right shows pitting and corrosion of a stainless steel surface. This was caused by a biofilm, whose presence influenced how and how fast minerals were deposited on the surface. This, in turn, modified the electrochemical properties of the stainless steel, which caused the pitting corrosion of this seemingly impervious metal.
Can we extend what we learn about this kind of biofilm to other sorts of biofilms, such as plaque on teeth? Apparently so, as discussed in the rest of this section.
Here are some of the more evident characteristics common to all observed biofilms:
- biofilms are dynamic and responsive to their environment; that is, they can adapt to changes in their environment.
- a phenomenon known as detachment seems to be common among all biofilms. Bacterial cells can detach from their biofilm colony individually or in clumps.
- when individual microorganisms detach from a biofilm, these isolated microorganisms are relatively easy to kill with chemicals designed for this purpose.
- when microorganisms detach from their biofilm colony in clumps, the clumps are pieces of the biofilm that are at the moment not attached to a surface; in this case they maintain the protective properties of the original biofilm and are thus much more difficult to kill.
- in the right conditions, biofilms can migrate across surfaces over a period of time in a variety of ways, as illustrated below.
Biofilms appear to show aspects of both solids and liquids—much like slug slime—and fall into a category called "viscoelastic." However, as biofilms collect sediment, or become scaled with rust or calcium deposits, they become less fluid and more like a brittle solid.

2. Genetic expression is different in biofilm bacteria when compared to planktonic bacteria
Here is a somewhat startling characteristic of bacteria in a biofilm as observed by biofilm scientists and engineers. The same kind of bacteria are different when they are in a biofilm than when they are isolated in planktonic form (that is, floating as single cells in water). Let's think about this for a moment. This is one of those scientific discoveries that seems counterintuitive. It might seem so obvious that a bacteria cell is a bacteria cell is a bacteria cell that one might not even think to check whether a particular bacterium is different when it is found in different environments.
The details of how this is determined is an advanced topic, but you might find it interesting to hear how it is done.
The double-stranded helix structure of molecular DNA (deoxyribonucleic acid), discovered in 1953 (by Watson and Crick), has become a familiar image. DNA molecules, composed of units called genes, carry the "instructions" that determine characteristics of living organisms and comprise the genetic material passed along to offspring through reproduction.
The genes that form DNA molecules also play a crucial role in cellular activities. Simple cells like bacteria control their internal functions using various parts of their genetic code to initiate chemical activities. So, for instance, consuming nutrients and getting rid of waste products are processes that are carried out under the influence of genetic instructions. When genes are activated to make chemical products (amino acids and proteins), they are said to be upregulated; when the genes are de-activated, they are downregulated. The proteins made by activated genes constitute about half of the material inside a cell, and are responsible for numerous activities that keep a cell viable.
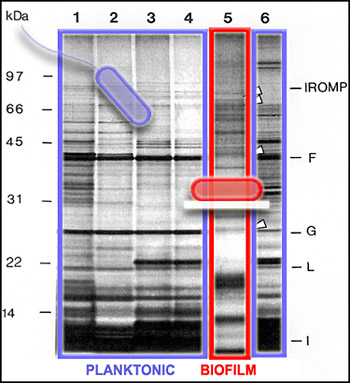
Since not all of the genes in a cell are activated to make proteins all of the time, we can get a picture of cellular activity by examining the proteins produced by cells at a particular time. One way to get this kind of protein "snapshot" is by a technique called SDS-PAGE (for "Sodium Dodecyl Sulfate" and "PolyAcrylamide Gel Electrophoresis"). This technique allows scientists to see large (nearer the top) and small (nearer the bottom) cellular proteins as dark bands in an array of columns. In the SDS PAGE gel above, we see proteins from the outer membranes of planktonic (outlined in blue, Lanes 1-4 and 6) and biofilm (outlined in red, Lane 5) bacteria, of a single strain. The bands of proteins are strikingly different, telling us that the planktonic and biofilm forms of a single species are expressing different genes, and therefore carrying out different activities.
So what? Beyond the intellectual interest this holds for biofilm scientists and engineers, what practical use does this knowledge have? One example is in the development of antibiotics. These drugs traditionally have been developed to kill planktonic bacteria under the assumption that they would kill the same bacteria wherever they were found. We now know, however, that
- Planktonic bacteria are more susceptible to antimicrobial chemicals designed to kill them than are biofilm bacteria, and
- Many of the infections plaguing humans are actually caused by bacteria in the biofilm mode of growth, not the planktonic mode of growth.
Put these two things together with the fact that traditional antibiotics have been designed for and tested on bacterial cells in their relatively unprotected, planktonic state and we can begin to understand why it is that antibiotics don't work well on these same bacteria when they exist in a biofilm—the same bacterium is different in the biofilm state than in the planktonic state for which the antibiotic was designed and tested!
This presents scientists and engineers with a new challenge, namely the development of new classes of antibiotics that target bacteria that exist in the biofilm state. Understanding the genetic activity of biofilm bacteria will help us to find new ways to target these cells and disrupt their functions.
3. Biofilm cells can coordinate behavior via intercellular "communication" using biochemical signaling molecules
Another characteristic of cells found in a biofilm is that they can communicate with each other. Really, in order for any community to succeed, there must be good communication among its members. Biofilm communities appear to be no different. Now how, you might ask, can single-cell microorganisms, such as bacteria, communicate with each other? One of the fascinating aspects of bacterial community living is that it provides a setting for bacteria to communicate using chemical signals. There is evidence that some of these chemical signals, produced by cells and passed through their outer membranes, may be interpreted not just by members of the same cell species, but by other microbial species that are part of the same biofilm community — and perhaps even by more complex organisms in some cases. The sensing of these chemical signals by neighboring cells in the biofilm can cause the neighboring cells to behave differently. How? By causing different genetic expression to occur in those cells, as described in the account in subsection 2 above.
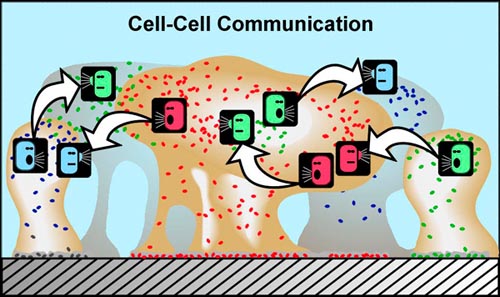
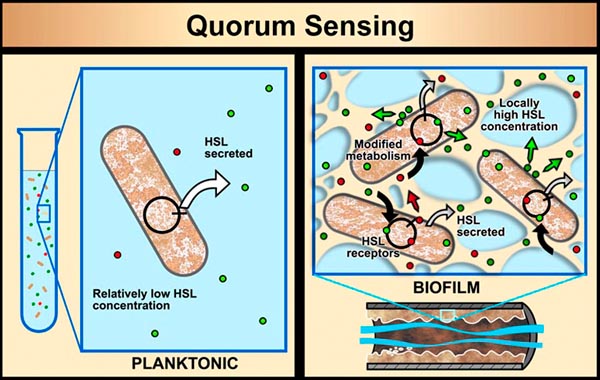
In planktonic populations of these same kinds of cells, chemical signals produced by the cells are simply not concentrated enough when passed through the water to cause changes in genetic expression. However, in biofilms, the matrix (glue) material (EPS) that holds cells close together allows concentrations of cell-produced chemical signal molecules to build up in sufficient quantity to cause changes in cellular behavior. Bacterial populations will activate some genes only when they are able to sense, via cell signaling, that their population is numerous enough to make it advantageous and/or "safe" to initiate that genetic activity.
For example, some bacterial pathogens (the bad-guy, disease-causing bacteria) will not produce toxins until they sense that an adequate population of themselves has been established to survive host defenses (e.g., antibodies, produced by a host human or animal, that can kill the bacteria). This system of population recognition has been termed "quorum sensing" (you've got it right; this comes from the same term used in a committee when enough members are present to legally take some action). It was first observed in the marine bacterium Vibrio fischeri, which can produce light after a sufficient population of this bacterium has developed.
The discovery that simple cells are capable of coordinated behavior has given us an entire, new appreciation of their survival strategies. There is also good evidence that cell signaling can cause cells of the same variety to form sub-populations that carry out different activities. For example, in the late 1990s an investigation of a biofilm community the marine bacterium Pseudoalteromonas revealed two physiologically distinct subpopulations. In effect there was a cellular division of labor: one group stayed attached to the surface and made nutrient available to the the second group, which reproduced and released daughter cells to the surrounding water.
In summary, the life of a simple, single-cell microorganism, such as a bacterium, is not so simple after all! And when these microorganisms are found in a biofilm colony their complexity increases tremendously. In order to treat and/or make beneficial use of biofilms, we must continue to identify and exploit the characteristics that are exhibited by microorganisms that form a biofilm.
4. Biofilms are less susceptible to antimicrobial agents
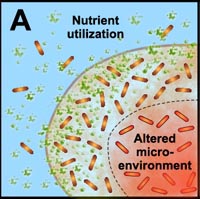
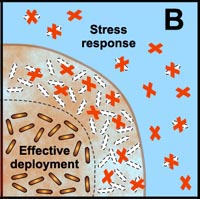
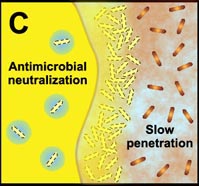
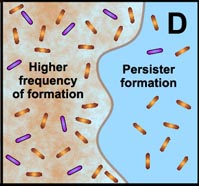
Another final characteristic of biofilms that we explore in this section—one that we have hinted at numerous times in this module—is that the microorganisms in a biofilm are much less susceptible to antimicrobial agents (chemicals designed to kill those microorganisms) than are the same microorganisms found in a planktonic state. Many studies have shown that the multicellular construction of biofilms affords protection for the cells that are part of these biofilms. This protection is the result of intrinsic shifts in the way these cells behave (through different genetic expression as described in subsection 2 above) once they attach to surfaces and begin to form biofilms. Some of the hypothesized mechanisms of protection from antimicrobial agents are pictured in the diagram below.
A. Free-floating cells utilize nutrients, but do not have sufficient metabolic activity to deplete substrates from the neighborhood of the cells. In contrast, the collective metabolic activity of groups of cells in the biofilm leads to substrate concentration gradients and localized chemical microenvironments. Reduced metabolic activity may result in less susceptibility to antimicrobials. |
B. Free-floating cells carry the genetic code for numerous protective stress responses. Planktonic cells, however, are readily overwhelmed by a strong antimicrobial challenge. These cells die before stress responses can be activated. In contrast, stress responses are effectively implemented in some of the cells in a biofilm at the expense of other cells which are sacrificed. |
 |
 |
 |
 |
C. Free-floating cells neutralize the antimicrobial agent. The capacity of a lone cell, however, is insufficient to draw down the antimicrobial concentration in the neighborhood of the cell. In contrast, the collective neutralizing power of groups of cells leads to slow or incomplete penetration of the antimicrobial in the biofilm. |
D. Free-floating cells spawn protected persister cells. But under permissive growth conditions in a planktonic culture, persisters rapidly revert to a susceptible state. In contrast, persister cells accumulate in biofilms because they revert less readily and are physically retained by the biofilm matrix. |


