What are key characteristics of biofilms?
Biofilms are complex, dynamic structures
Bacterial biofilms are remarkably heterogeneous in virtually all parameters that can be measured accurately and reproducibly. These heterogeneities: structural, physiological, chemical, ecological, electrical, etc., have been implicated as the cause of many phenomena characteristic of the attached—as opposed to the planktonic—mode of growth. Pure cultures are virtually non-existent in the world outside the laboratory. Microorganisms, like other organisms, exist in communities where a variety of interactions exist.
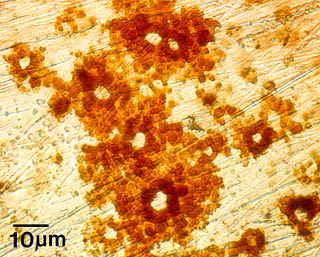
One example of the effects of microbial colonization on metal surfaces is Microbially Influenced Corrosion (MIC). The presence of microorganisms modifies deposition and dissolution rates of minerals, and by this mechanism, influences the electrochemical properties of the metals or alloys. Pitting corrosion, as seen at right in this example from a stainless steel surface, results from this activity.
Biofilms are also dynamic and responsive to their environment. Bacterial cells may detach from biofilms individually or in clumps. When they detach in clumps, they retain the reduced susceptibility to antimicrobials characteristic of biofilms. In the right conditions, biofilms can also migrate across surfaces over a period of time in a variety of ways, as illustrated below.
Biofilms appear to show aspects of both solids and liquids—much like slug slime—and fall into a category called "viscoelastic." However, as biofilms collect sediment, or become scaled with rust or calcium deposits, they become less fluid and more like a brittle solid.

Genetic expression is different in biofilm bacteria when compared to planktonic bacteria
The double-stranded helix structure of molecular DNA (deoxyribonucleic acid), discovered in 1953, has by now become a familiar image. The genes of all living organisms, composed of DNA, carry the “instructions” that determine the characteristics (phenotype) of the organism and are also the genetic material that is transmitted to the next generation by sexual or asexual reproduction.
These genetic instructions function primarily through the synthesis of proteins. Some of these are structural components of the cell (flagellae for example), and many are enzymes the biological catalysts that carry out chemical reactions in the cell. These enzymes are responsible for energy production, acquiring nutrients from the surrounding environment, waste disposal and the manufacturer of the various components of which the cells are made. What may be surprising is that not all genes in the cell function all the time. There are to be sure certail “housekeeping” genes whose function is so essential that they function all the time, but other genes such as those used in spore production are in use only during certain points in the bacterial life cycle. So genes may be “upregulated” (turned on) or down regulated (turned off) as required by the cell. This ability to regulate gene function, like turning off a light switch as you leave a room, results in a significant savings in energy.
Interestingly, it has been discovered that within minutes of when a bacterium attaches to a surface it undergoes a remarkable change in gene function. Many operating genes (e.g. flagellae) are down regulated while others (slime production) are upregulated. How do we know?
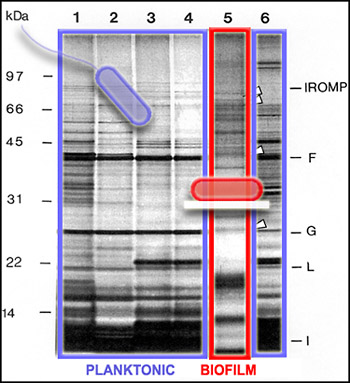
We can get a picture of cellular activity by examining the proteins produced by cells at a particular time. One way to get this kind of protein "snapshot" is by a technique called SDS-PAGE (for "Sodium Dodecyl Sulfate" and "PolyAcrylamide Gel Electrophoresis"). This technique allows scientists to see large (nearer the top) and small (nearer the bottom) cellular proteins as dark bands in an array of columns. In the SDS PAGE gel above, we see proteins from the outer membranes of planktonic (outlined in blue, Lanes 1-4 and 6) and biofilm (outlined in red, Lane 5) bacteria, of a single strain. Although we cannot tell from this gel just what these proteins are doing, we can see that the bands of proteins are strikingly different, telling us that the planktonic and biofilm forms of a single species are expressing different genes, and therefore carrying out different activities.
Estimates vary, but some suggest that as much as 40% of the genes of a bacterium may undergo up or down regulation in the transition from the planktonic to the biofilm state. So, even though the genes present haven’t changed at all the expression of those genes has undergone a dramatic shift, it is almost as if the two states (planktonic and biofilm) were entirely different organisms.
What are the implications of this difference in genetic expression? One example is in the development of antibiotics. These drugs have traditionally been developed to kill planktonic bacteria. We now know, however, that planktonic bacteria are more susceptible to antimicrobials than biofilm bacteria—and also that many of the infections plaguing humans are actually caused by bacteria in the biofilm mode of growth. So traditional antibiotics have been targeting bacterial cells in their relatively unprotected state. We will need to develop new classes of antibiotics that target bacteria without in the biofilm state. Understanding the genetic activity of biofilm bacteria will help us to find new ways to target these cells and disrupt their functions
Biofilm cells can coordinate behavior via intercellular "communication" using biochemical signaling molecules
One of the fascinating aspects of bacterial community living is that it provides a setting for bacteria to communicate using chemical signals. There is evidence that some of these chemical signals, produced by cells and passed through their outer membranes, may be interpreted not just by members of the same species, but by other microbial species—and perhaps even by more complex organisms in some cases.
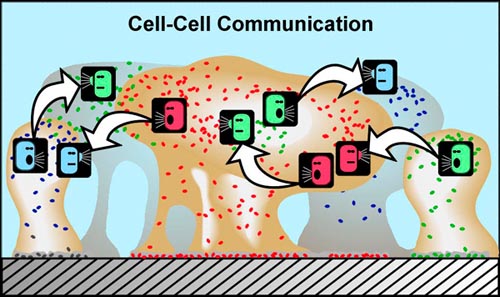
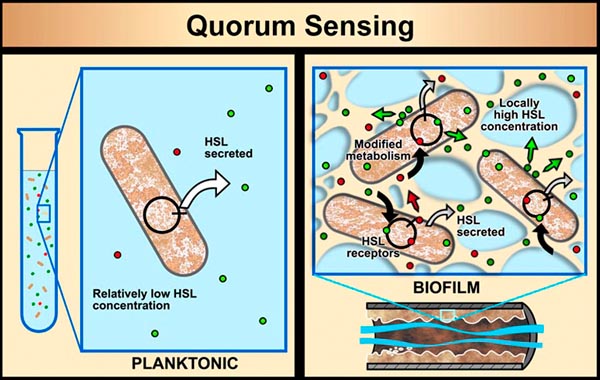
In planktonic populations, chemical signals produced by cells are not concentrated enough to cause changes in genetic expression. However, in biofilms, the matrix material (EPS) that holds cells in close proximity allows concentrations of signal molecules to build up in sufficient quantity to effect changes in cellular behavior. Bacterial populations will activate some genes only when they are able to sense, via cell signaling, that their population is numerous enough to make it advantageous and/or "safe" to initiate that genetic activity. For example, some bacterial pathogens will not produce toxins until they sense that an adequate population has been established to survive host defenses. This system of population recognition has been termed "quorum sensing." It was first observed in the marine bacterium Vibrio fischeri, which can produce light after a sufficient population has developed.
The discovery that simple cells are capable of coordinated behavior has given us an entirely, new appreciation of their survival strategies. There is also good evidence that cell signaling can cause cells of the same variety to form sub-populations that carry out different activities. For example, in the late 1990s an investigation of a biofilm community the marine bacterium Pseudoalteromonas revealed two physiologically distinct subpopulations. In effect there was a cellular division of labor: one group stayed attached to the surface and made nutrient available to the the second group, which reproduced and released daughter cells to the surrounding water.
Biofilms are less susceptible to antimicrobial agents
Many studies have shown that the multicellular construction of biofilms affords protection for cells. This protection is the result of intrinsic shifts in genetic expression when floating bacterial cells attach to surfaces and begin to form biofilms. Some of the hypothesized mechanisms of protection from antimicrobial agents are pictured in the diagram below.
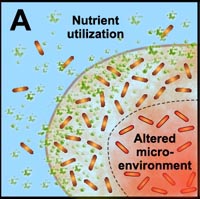
A. Free-floating cells utilize nutrients, but do not have sufficient metabolic activity to deplete substrates from the neighborhood of the cells. In contrast, the collective metabolic activity of groups of cells in the biofilm leads to substrate concentration gradients and localized chemical microenvironments. Reduced metabolic activity may result in less susceptibility to antimicrobials. |
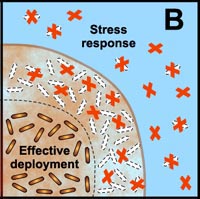
B. Free-floating cells carry the genetic code for numerous protective stress responses. Planktonic cells, however, are readily overwhelmed by a strong antimicrobial challenge. These cells die before stress responses can be activated. In contrast, stress responses are effectively implemented in some of the cells in a biofilm at the expense of other cells which are sacrificed. |
 |
 |
 |
 |
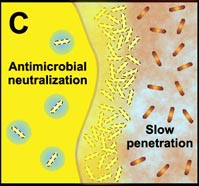
C. Free-floating cells neutralize the antimicrobial agent. The capacity of a lone cell, however, is insufficient to draw down the antimicrobial concentration in the neighborhood of the cell. In contrast, the collective neutralizing power of groups of cells leads to slow or incomplete penetration of the antimicrobial in the biofilm. |
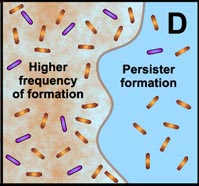
D. Free-floating cells spawn protected persister cells. But under permissive growth conditions in a planktonic culture, persisters rapidly revert to a susceptible state. In contrast, persister cells accumulate in biofilms because they revert less readily and are physically retained by the biofilm matrix. |


