What is the evidence that biofilm forms in chronic wounds?
Molecular analysis
Culture based methodologies for the identification and quantification of microorganisms form the basis of most clinical lab studies. However, many microorganisms have been overlooked in these studies due to the inability to grow them in culture. Denaturing gradient gel electrophoresis (DGGE) of PCR-amplified 16S rRNA genes was used to characterize biofilm communities from wound samples provided by the Southwest Regional Wound Care Center. DGGE is a method for separating similar sized DNA fragments based on sequence. Bacterial community diversity (i.e. number of different species) can be estimated from the number of bands in gels from each species.
Tissue samples were obtained during standard sharp debridement of wounds during normal wound care management. Each sample was homogenized prior to extraction. Total DNA was extracted using the Bio101 FAST DNA Spin Kit for soil (QBioGene). Extracted total DNA was stored as -20º C prior to shipment to CBE-MSU for PCR and DGGE. The DNA sent to MSU-CBE was used as a template for polymerase chain reaction (PCR).
Bacterial DNA from the wound samples was initially amplified using two sets of universal Eubacterial forward and reverse primers containing a GC clamp. These primer sets included: 1070F (5’ ATG GCT GTC GTC AGC T 3’) combined with 1392R + GC Clamp (5’ CGC CCG CCG CGC CCC GCG CCC GGC CCG CCG CCC CCG CCC C ACG GGC GGR GRG TAC 3’) and 518R + GC Clamp (5'GTA TTA CCG CGG CTG CTG G 3') combined with a 357F (5" CGC CCG CCG CGC CCC GCG CCC GGC CCG CCGC CCC CGC CCC C CTA CGG GAG GCA GCA G 3') (Integrated DNA Technologies). Primer reactions and DNA amplification were performed using a PTC-100 Programmable Thermal Controller (MJ Research). Verification of the presence of DNA was assessed in 1.5% agarose gels.
Amplified DNA was separated by DGGE using a 40% to 70% denaturing gradient in 8% to 12% polyacrylamide gels following recommended manufacturer protocols (BioRad). DGGE can detect differences in single base changes anywhere along the strand of amplified DNA. The double stranded, amplified DNA is subjected to an increasing gradient of denaturant during gel electrophoresis. The DNA strands then melts, creating a branched molecule which stops its migration within the gel. The temperature at which the DNA melts (TM) is a function of the sequence of the DNA molecule.
In initial studies, the combination of 518R + GC Clamp and 375F primers resulted in the highest number of bands and was selected for subsequent amplifications. DGGE results for a number of wound samples are shown in Figure 9. Each wound sample displayed a unique banding pattern within the DGGE gels, although many samples contained bands at similar locations within the gels. Similar findings have been reported from DGGE of samples from venous leg ulcers (Davies et. al., 2004). In contrast to the venous leg ulcer study, an identical band was not present in all of the samples.
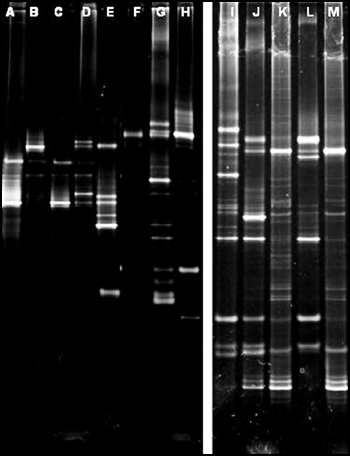
Information on wound types and culture results for the above DGGE gels are presented in Table 1. In one case, (A) no microorganisms were cultured, but a distinct band was apparent after PCR and DGGE of extracted DNA from the chronic wound sample. In many cases, one to three organisms were cultured from the wound samples but many bands are present in the DGGE analysis indicating that multiple species were present. These findings underscore the utility of molecular methods for detecting unculturable microorganisms. These preliminary studies also suggest that the samples from the diabetic foot ulcers had the most species diversity (i.e. bands) and venous leg ulcers had the least species diversity. However, a detailed analysis of more samples will be necessary to confirm this suspicion.
| Gel Lane* | Wound type | Genera/species cultured |
A |
Non-healing surgical wound |
Enterobacter, Pseudomonas |
B |
Venous leg ulcer |
None |
C |
Calciphylaxis |
Pseudomonas, Staphylococcus |
D |
Venous leg ulcer |
Escherichia coli, Staphylococcus aureus |
E |
Diabetic foot ulcer |
Streptococcus (Group B), Citrobacter, Staphylococcus |
F |
Venous leg ulcer |
Pseudomonas, Staphylococcus |
G |
Chronic wound |
Enterococcus (Group B), Escherichia coli, Staphylococcus aureus |
H |
Venous leg ulcer |
Pseudomonas |
I |
Diabetic foot ulcer |
Enterococcus (Group D) |
J |
Diabetic foot ulcer |
Citrobacter freundii, Staphylococcus aureus |
K |
Decubitus ulcer |
Staphylococcus aureus |
L |
Diabetic foot ulcer |
Staphylococcus |
M |
Non-healing surgical wound |
Pseudomonas, Staphylococcus |
Overall, our preliminary microbiological studies of wounds indicated that biofilms were prevalent in chronic wounds and rare in acute wounds. These biofilms were polymicrobial communities, and each wound sample appeared to have unique community structure. Although the mere presence of complex microbial biofilms in wounds does not provide evidence that they are a barrier to healing, clinical observations of chronic wounds are consistent with those of other biofilm-related diseases and further research is warranted to assess the role of biofilms in the prevention of wound healing.
